RNAi
In the 1990s, a small industry-focused research team at CSIRO Plant Industry in Canberra, led by Peter Waterhouse, was investigating how potatoes and cereals could be protected against viruses. One half of the team, which included Michael Graham, Neil Smith, and Geoff Ellacott, were working on potatoes. The other half, Ming-Bo Wang, David Abbott and Limin Wu, were working on barley and rice. During this research, they discovered something spectacular – how to activate one of the most fundamental gene control pathways in plants. The pathway, which is now referred to as RNA interference (RNAi), is guided by double-stranded RNA and works by degrading target single stranded RNAs.
While the CSIRO teams, were making their discovery in plants, a similar breakthrough was being made in the genetic model nematode (Caenorhabditis elegans) by the US researchers Andrew Fire and Craig Mello. Together these discoveries have led to the development of gene silencing technologies that have revolutionised the way in which genes and developmental control in plants and animals are studied, and the generation of numerous biotechnology applications. The CSIRO technologies use ‘hairpin’ RNA to induce and guide gene silencing and this approach has formed the basis for more than 100 patent applications and the production of gene silencing plasmid vectors that have been adopted by over 3 500 public or private-funded research laboratories around the world.
For their work, Andrew Fire and Craig Mello were awarded the 2006 Nobel Prize in Medicine and Physiology, and Peter Waterhouse and Ming-Bo Wang were awarded the 2007 Prime Minister’s Prize for Science. The CSIRO team was also awarded the CSIRO Chairman’s Medal in 2005.
The early studies on coat protein mediated virus protection
The introduction of recombinant DNA techniques and the development of gene transfer and plant regeneration procedures in the 1980s facilitated the transfer of genes not only between plants of different species but also the transfer of genes from viruses, bacteria, fungi and animals into plants. These transferred genes are called ‘transgenes’, the process of introducing them into plants is termed ‘transformation’ and the ability to transform plants with transgenes has provided unparalleled opportunities for crop improvement.
In 1986, the American plant virologist Roger Beachy and his colleagues showed that tobacco plants expressing a transgene derived from the coat protein gene of tobacco mosaic virus (TMV) were highly resistant to infection by TMV. This finding motivated major research activity around the world and led to many published reports of protection against a range of viruses in a spectrum of plant species.
All was not exactly what it seemed
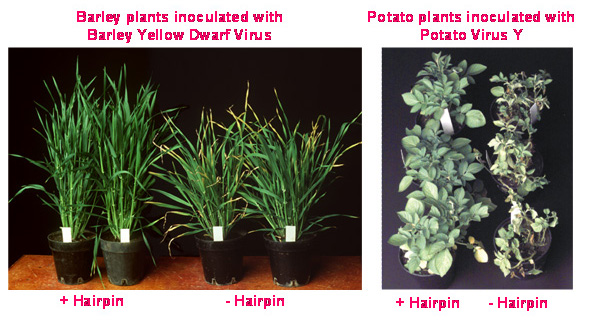
However, when some of these transgenic plants were examined at the molecular level, the results were rather surprising. In Roger Beachy’s plants, the more transgenic TMV coat protein (or messenger RNA coding for the protein) that was expressed, the stronger was their resistance to infection by TMV. However researchers, using transgenes made from other viruses, often did not see the same correlation between transgene expression level and degree of virus resistance. Indeed, when the CSIRO team, inspired by Roger’s results, set out to protect potatoes (and tobacco – as a test plant which is easier to transform) from potato leaf roll virus (PLRV) and potato virus Y (PVY) and cereals from barley yellow dwarf virus (BYDV), they too found this puzzling lack of correlation. For example, out of a batch of more than 30 independent transgenic potato plants transformed with a PLRV coat protein gene, a few had good PLRV resistance but they also had low levels of transgene messenger RNA and undetectable levels of PLRV coat protein. Other plants from the same batch had higher levels of transgene expression but were completely susceptible to PLRV infection.

Ironically, it was Roger Beachy who pointed the CSIRO team towards the involvement of RNA, not protein, in the protection against virus infection. At a small plant molecular biology conference in Cairns, Australia, Peter Waterhouse was discussing these perplexing results with Roger, who suggested that he look at a research paper just published by two US researchers, John Lindbo and Bill Dougherty. These researchers had shown that transforming plants with transgenes designed to express RNAs of viral origin, but which could not be translated into proteins, could still activate a mechanism that inhibited infection by the virus. They also showed that the mechanism operated by specifically degrading both the mRNA from the introduced transgene and the genomic RNA of the inoculated virus, and that this process had great similarities with a phenomenon called post-transcriptional gene silencing (PTGS) – which had been recently described using transgenes of non-viral origin.
CSIRO Plant Industry looks more deeply
Bill Dougherty’s US group had shown that an important attribute of the virus-resistant transgenic plants was that they contained multiple (often more than 5) copies of the virus-derived transgene. From this they proposed a ‘threshold’ model: if a cell expresses molecules of the same RNA sequence at a level higher than a certain threshold, this induces a mechanism that specifically destroys that RNA. If the highly expressed transgene RNA that triggers this mechanism contains viral sequence, then this will also cause degradation of the RNA genome of a virus containing this sequence. This model inspired the CSIRO team to make DNA constructs aimed at increasing the copy number of virus-derived transgenes delivered into each plant. To do this they made various transformation constructs that contained two copies of the virus-derived transgene in the same piece of DNA.
Construct version-1 contained two sense copies, version-2 contained two antisense copies and version-3 contained both one sense and one antisense copy. Challenging plants which had been transformed with these constructs gave an unexpected, but exciting, result.
The proportion of plants showing good virus protection was much higher in those transformed with version-3 than those transformed with either of the other two versions. Furthermore, some of the version-3 protected plants had only one piece of the transferred DNA, i.e. only one sense and one antisense gene. This led the CSIRO team to think that it was the production of dsRNA that was causing this effect, not the number of gene copies.

It’s the double stranded RNA
While doing these double transgene experiments, the CSIRO team had also been repeating the Dougherty ‘no-protein’ transgene approach. They made a transgene from PVY into which they had engineered a ‘premature stop’ codon, to prevent the expression of the viral protein, and from this made both sense and an antisense gene constructs. Just as Bill Dougherty and his colleagues had predicted: using either construct, only the tobacco plants which were, by chance, transformed with multiple copies showed good virus resistance. A by-product of these experiments was the generation of many independent transgenic tobacco plant lines that had only a single sense or a single antisense PVY transgene – all of them susceptible to PVY infection. These plants were the perfect resource to confirm the dsRNA model.
To scientifically prove whether the high level of virus resistance obtained with the CSIRO version-3 construct was due to its structure or to its production of dsRNA, the CSIRO researchers crossed a plant containing only one premature stop codon PVY sense transgene with a plant containing only one premature stop codon antisense gene and then tested the progeny for virus resistance. The sense and antisense genes were completely unlinked so combining them in a progeny plant would not give an inverted-repeat transgene structure but would produce sense and antisense RNA that could hybridise to form dsRNA.
The results were conclusive and consistent with the dsRNA model. One quarter of the progeny plants were completely resistant to the virus and three quarters of them were susceptible, and when the genetic make-up of these plants was examined, the answer was there. One quarter of the plants had inherited only the sense gene, one quarter had inherited only in the antisense gene, and one quarter had inherited neither gene; all of these plants were susceptible to the virus. However, every plant that belonged to the quarter which had inherited both the sense and the antisense gene was completely protected against to the virus. This experiment was repeated with five different sense lines crossed with five different antisense lines and in every case, the progeny plants inheriting both the sense and antisense genes were virus resistant. This told the CSIRO team that dsRNA containing viral sequences from the transgenes was triggering and directing the destruction of these sequences in the single stranded RNA of the challenging virus. There were two further insights: (i) if this could work for the degradation of viruses, it could also work for the degradation of messenger RNA of any endogenous gene; (ii) there is probably no need to make two separate RNAs which hybridise into dsRNA, a transgene expressing RNA that folds back and hybridises with itself (to form a structure like a hairpin) would also work.
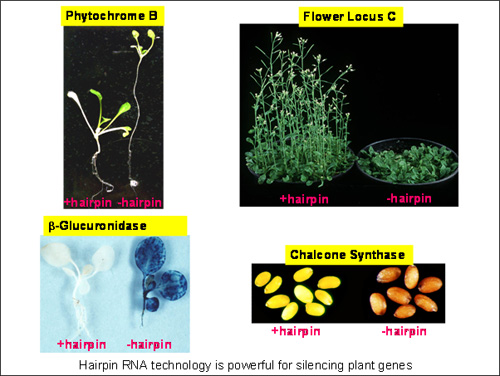
They made a hairpin transgene containing sequences derived from the blue’ reporter gene, beta-glucuronidase (GUS), and engineered rice cells to contain various permutations of a GUS gene and a GUS-derived hairpin (hp) gene. When the different cell lines were stained to show GUS expression, the cells with the GUS gene alone gave a strong blue colour, but those with both the GUS gene and the GUS-hp gene remained white. These findings provided the first proof that gene silencing in plants can be induced by double-stranded RNA or hairpin RNA and were published in a historic paper in the Proceedings of the National Academy of Science, USA on the 10 November 1998 (Waterhouse et al., 95: 13959-13964).
Meanwhile on the other side of the world
Meanwhile the American molecular geneticists Andrew Fire, at the Carnegie Institution of Washington in Baltimore, and Craig Mello, at the University of Massachusetts, were studying a somewhat similar phenomenon using the nematode Caenorhabditis elegans as their genetic model. They used expression of the unc-22 gene as their genetic target. unc22 encodes an abundant but nonessential myofilament protein. Several thousand copies of unc-22 mRNA are present in each striated muscle cell and a link had been described between unc-22 levels and the phenotype of the organism.
Like the plant geneticists at CSIRO, they found, to their surprise, that double-stranded RNA was substantially more effective at producing interference of unc-22 gene function than was either strand individually. After injection into adult animals, purified single strands had at most a modest effect, whereas double-stranded mixtures caused potent and specific interference. The effects of this interference were evident in both the injected animals and their progeny. Only a few molecules of injected double-stranded RNA were required per affected cell, arguing against stoichiometric interference with endogenous mRNA and suggesting that there could be a catalytic or amplification component in the interference process. These findings were published in the 19 February issue of Nature, (Fire et al., 1998, 391: 806-811), nine months before the Canberra group.
Common mechanisms in plants and animals
These studies revealed that both plants and animals recognise double-stranded RNA or hairpin RNA as foreign, break it up and use the broken-up pieces as a guide to attack any RNA in the cell with the same gene sequence. By artificially introducing double-stranded RNA or hairpin RNA into plants, the two teams were in effect tricking a cell into believing that the messenger RNA of the gene they wanted to silence was foreign and dangerous and therefore needed to be destroyed.
In their paper in Nature, Fire et al. comment that they did not yet know the mechanism of RNA-mediated interference in C. elegans but offered some observations that add to the debate about possible targets and mechanisms.
First, dsRNA segments corresponding to various intron and promoter sequences did not produce detectable interference. Although consistent with interference at a post-transcriptional level, these experiments do not rule out interference at the level of the gene.
Second, they found that injection of dsRNA produced a pronounced decrease or elimination of the endogenous mRNA transcript.
Third, dsRNA-mediated interference showed a surprising ability to cross cellular boundaries. Injection of dsRNA into the body cavity of the head or tail of C.elegans produced a specific and robust interference with gene expression in the progeny brood and dsRNA injected into the body cavity or gonad of young adults also produced gene-specific interference in somatic tissues of the injected animal.
The Canberra group in their paper in PNAS – described by Nobel Laureate, Phillip Sharp as ‘The purest demonstration that dsRNA mediates gene silencing in plants’, (Sharp PA, Genes Dev, 1999, 13: 139-410) – postulated a model for how RNA interference was occurring. The cornerstone of their model was that gene silencing is induced by dsRNA and that this is mediated by an RNA-dependent RNA polymerase, present in the cytoplasm, that requires a dsRNA template. This specificity for a dsRNA template is the key to producing sequence-specific degradation. The dsRNA is formed by hybridisation of complementary transgene mRNAs or complementary regions of the same transcript. The dsRNA-dependent RNA polymerase produces complementary RNA (cRNA) to which degradative RNase molecules are attached. These cRNA-RNase molecules hybridise to the endogene mRNA or viral RNA and cleave the single-stranded RNA adjacent to the hybrid. The cleaved single-stranded RNAs are further degraded by other host RNase enzymes. If the transgene is derived from a viral sequence, this will provide sequence-specific cleavage of viral genomic and subgenomic RNAs, thus preventing viral infection. If the transgene is derived from one of the plants own genes (endogene), this will lead to cleavage and degradation of the endogene mRNA and consequent loss of function.
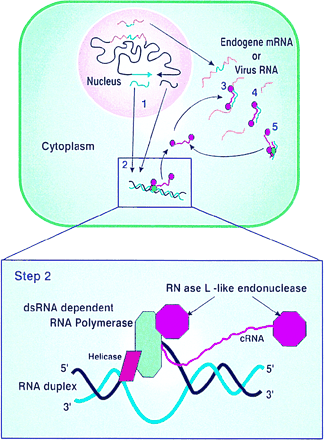
It was subsequently shown that, in both plants and animals, dsRNA or hairpin RNA is cleaved into small RNAs by an enzyme called Dicer and one of these strands is transferred to a nuclease called Argonaute which it uses to recognise and then degrade ssRNA. It is also known that these small RNAs are amplified by an RNA dependent RNA polymerase. Thus, not quite all, but most of the concepts of the CSIRO model were correct.
Hairpin RNA gene silencing
The discovery that double-stranded RNA can be used by the plant cell to degrade viral and plant RNAs led to the development of a series of technologies by CSIRO based on hairpin RNA-induced gene silencing. Their hairpin RNA constructs for gene silencing and their construct design principles, including the use of intron spacers and high throughput recombinase cloning, are now widely adopted for gene silencing in plants and animals and form the basis of the AGRIKOLA, European Framework 5 project aimed at silencing every gene in the Arabidopsis genome.
The gene silencing technology is supported by more than 100 patent applications and has been adopted by over 3 500 public or private-funded research laboratories around the world. It has become a powerful tool in investigating gene functions and in improving agronomical traits in plants.
These technologies can be used to stop the expression of any plant or virus gene by destroying their RNA message that directs protein production in a cell. Selective silencing of a specific gene, such as a gene for a particular colour in flowers or a specific fatty acid in oils, can result in altered traits in the plant. Such selective gene silencing, using the CSIRO-developed technologies, has been successfully applied to producing plants that are resistant to viruses and insects, produce healthy foods and therapeutic antibodies, or exhibit highly valuable commercial traits.
The science has been exciting
At one point we seemed to spend more time talking law then we did talking science
The conducive environment that led to the success
The discovery of double-stranded RNA-induced gene silencing by the CSIRO team has been a good example of how sustained research efforts, collaborative research environments, and team work can yield ultimate rewards. The work to understand the gene silencing-based antiviral defence in plants was continued uninterrupted for many years thanks to the strong support from CSIRO Plant Industry management.
Close collaborations and frequent exchange of ideas within the team and with colleagues working on plant development, barley improvement, cereal transformation, oil modification, and various other aspects of molecular biology and virology in CSIRO Plant Industry, were critical for the initial breakthrough and especially for the subsequent development and validation of the CSIRO gene silencing technologies.
The discovery of double-stranded RNA-induced gene silencing in plants has come from incremental increases, involving a series of experiments with trial and error, and occasional moments of excitement and insight.
Besides Peter Waterhouse and Ming-Bo Wang, the dedication and perseverance by other members of the Canberra-based team have been critical in this discovery journey.
Honours and awards
For their discoveries on RNAi in plants Peter Waterhouse and Ming-Bo Wang were awarded the 2007 Prime Minister’s Prize for Science and the 2005 CSIRO Chairman’s Medal. For their discoveries on RNAi in animal cells (C. elegans), American scientists Andrew Fire and Craig Mello were awarded the Nobel Prize for Physiology or Medicine in 2006.
Sources
- Waterhouse PM, 2009, Personal communication.
- Wang MB, 2009, Personal communication.
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC, 1998, ‘Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans‘, Nature, 391: 806-811.
- Waterhouse PM, Graham MW, Wang MB, 1998, ‘Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA’, Proc. Natl Acad Sci, USA, 95: 13959-13964.
