Bacterial drug resistance, gene cassettes and integrons
From the mid 1980s, CSIRO researchers at North Ryde led by Ruth Hall, were involved in an effort to understand how antibiotic resistance genes get into and move about in bacterial populations rendering antibiotics ineffective for the treatment of bacterial diseases and hospital-acquired infections.
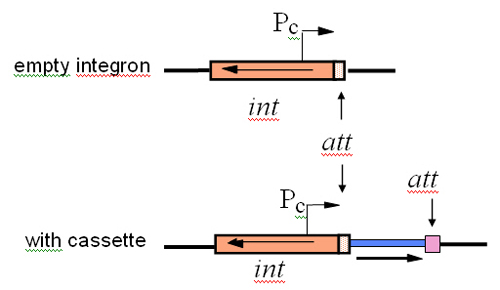
The work focused on a novel system for gene movement that they had discovered. This system involves small mobile elements, now called gene cassettes, that contain one (or occasionally two) gene(s), often antibiotic resistance genes, and a backbone element which was named an integron. The integron carries a site that the cassettes slot into and the gene for synthesis of an enzyme that slots them in.
Gene cassettes are now accepted to be an important source of new antibiotic resistance genes and, as multiple cassettes can reside together in an integron, providing resistance to more than one antibiotic. Integrons (with associated gene cassettes) have now been found in many different bacteria and represent an important way in which bacteria can add to the genes they carry.

Bacterial antibiotic resistance ‘ an ongoing problem
Antibiotic resistance is an ongoing and increasing problem that is reducing the efficacy of the relatively small collection of antibiotics that are available for treatment of infectious diseases caused by bacteria. As the incidence of resistance continues to rise in disease-causing bacteria, so does multiple resistance leaving few options for treatment. In fact, infections that are resistant to all or nearly all of the available antibiotics have emerged. At the same time, new antibiotics or more effective versions of existing ones are not being found. One way to approach this problem is to better understand what is leading to resistance, but at the time Ruth Hall began this work relatively little research funding was directed to this problem.
International research done in the 1960s and 1970s had revealed two important things about how antibiotic resistance genes are spread. First, it was found that there were small chromosomes called plasmids that replicate separately from the bacterial chromosome and can transfer or be transferred, often carrying antibiotic resistance genes with them, from the bacterium they were in to another one they encountered. This process, called horizontal gene transfer, is the way that resistance genes are shared amongst different types of bacteria.
The second discovery was that small defined segments of DNA called mobile elements, ‘jumping genes’ or transposons were found. These mobile elements could move themselves from one place in the DNA to another in the same or in a different DNA molecule within a single cell. Mobile elements provide another way for bacteria to take up new genes and also explain how resistance genes could have got onto the plasmids that are moving them into new bacteria. A lot of different transposons that carried genes conferring resistance to several different types of antibiotic were subsequently discovered. After this, it was generally thought that the story of gene movement was understood, and interest in working on resistance or funding such work waned.
Early work at CSIRO leads to a major surprise
In the mid 1980s, Ruth Hall, working at CSIRO’s Division of Molecular Biology at North Ryde, began what was expected to be a short project with Dr Dick Groot Obbink from Royal North Shore Hospital. The aim was to try to work out why probes they had made to enable them to screen for a gentamicin resistance gene causing problems at the hospital were lighting up strains that were resistant to quite different antibiotics. To do this, the gentamicin resistance gene that Dr Groot Obbink had cloned was sequenced in Hall’s laboratory. At that time, access to the earliest versions of the DNA sequence database (GenBank) was very limited and the Bioinformatics tools and computing power to search in it were not readily available.
In a happy coincidence, these things were being worked on by Dr Alex Reisner at CSIRO Molecular Biology and the Division owned a powerful VAX computer. Using this facility, Dr Hall was able to compare the sequence they had obtained to all other sequences known at the time. This revealed that the gentamicin resistance gene was surrounded by DNA sequences that were the same as ones located next to two quite unrelated genes that conferred resistance to streptomycin and to trimethoprim.
This finding explained the strange results that Dr Groot Obbink had seen, but was also a great surprise because it indicated that there was a way of putting different genes into the same place in a DNA molecule, and this hadn’t been seen before. Recognising the potential importance of this system, which provides an alternative to transposons for moving resistance genes into plasmids, Hall looked at the antibiotic resistance genes in another plasmid she was studying (for quite different reasons) at that time and the system was there too. In this plasmid, which had been isolated some 20 years previously in the UK, there was an oxacillin resistance gene in the cassette. Though it was by now clear that this system was being used a lot to acquire resistance genes, it had remained undetected largely because no one had looked further than transposons.
Defining and naming integrons and gene cassettes
The next phase in this project, in 1988, was to work out the details of how the system captures the resistance genes. This stage of the work was undertaken in collaboration with Dr Hatch Stokes at Macquarie University and eventually much of the work undertaken with Dr Stokes was funded by the NHMRC. The Division was now called Biotechnology and was to change again to Biomolecular Engineering (in 1990) and then to CSIRO Molecular Science (in 1997) before the project at CSIRO was terminated.
Initially, the regions surrounding further resistance genes that, from studying the literature, looked as if they might be part of the system were sequenced. A few other sequences derived in overseas labs also appeared at this time, and all of these sequences were used to search for the clues as to how it all worked. Sequence comparisons allowed the precise segments that are the gene cassettes to be defined and a variety of versions of distinctive sequence first seen on the other side of the gentamicin resistance gene were found next to each of the other resistance genes that had been found to be in a cassette. The analysis also revealed that a gene found in one of the conserved segments located next to the cassettes encoded a site-specific recombinase of a type known as integrases. Hence, the integron coded for a site-specific recombination enzyme and the incoming genes were associated with a short region of DNA that had distinctive features and presumably served as a site this enzyme could recognise. However, a new system needs a name – otherwise it’s very hard to talk about – and the name ‘integron’ was coined by Stokes and Hall in their landmark 1989 publication (Mol. Microbiol., 3: 1669-1683). The term ‘gene cassette’, by analogy with plug and play of audio cassettes, emerged a little later.
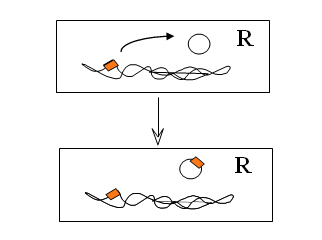
Functional demonstration of integration of gene cassettes in integrons
With the parts identified, experimental work could begin. Though Hall and Stokes were not the first to demonstrate that the recombinase (or integrase) was functional, they went on to demonstrate that the various recombination sites were active and that the one in the integron was very different to the ones associated with the cassettes.
Now, what they most wanted to achieve was to provide proof for the theory and demonstrate the integration of gene cassettes experimentally. In 1991, Dr Tina Collis joined the team at the North Ryde laboratory of CSIRO Biomolecular Engineering and in a short while she was able to use the integrase of the integron to first remove resistance gene cassettes and then to put them back into the correct site in an integron. This work showed that the site-specific recombinase could cause loss of gene cassettes, re-arrange them into a different order, and could integrate them into the site in the integron next to the integrase gene where they are normally found.
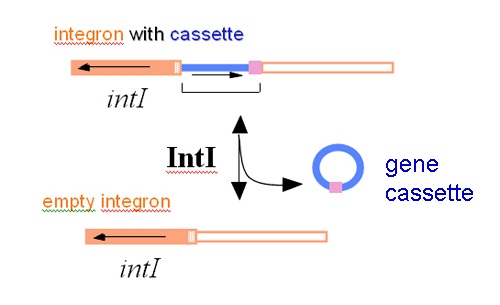
The resistance genes need a promoter if they are going to be expressed and there were none in the cassettes. Instead the promoter was found in the integron and in order to use it, the genes in the cassettes all face the same way. Dr Collis also demonstrated that the genes were expressed together from this promoter.
Many integrons, millions of gene cassettes
Another type of integron with a related but different integrase gene in it had been identified in Hall’s earliest work. This integron, which became class 2 with the original type as class 1, also carried cassettes with antibiotic resistance genes in them and eventually a third type was found in Japan. However, many of the cassettes that were found in the class 1 integrons didn’t carry antibiotic resistance genes and, very early on, it became obvious that the resistance gene story was simply the one seen first. Antibiotic resistant bacteria had been the source material for our studies and the use of antibiotics had provided a powerful selection to bring resistance genes into the now ubiquitous class 1 integrons.
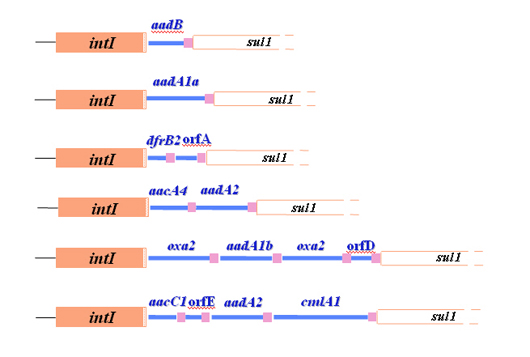
But, the cassettes with resistance genes in them must come from somewhere. Hence, it was exciting, though not a real surprise, when the first integron was subsequently found as part of a bacterial chromosome. Today hundreds of different integron integrases (and hence integron types) have been discovered and the number of cassettes seen so far indicates that there are millions of them, though only very rarely do they have any antibiotic resistance genes associated with them. Indeed, which bacteria the resistance genes come from and how the cassettes are created remain mysteries that scientists have yet to solve.
The situation today
Today, integrons and gene cassettes are an accepted part of the genetic repertoire of bacteria and not just for their role in the spread of antibiotic resistance. In gram negative bacteria, the incorporation of gene cassettes carrying new antibiotic resistance genes into class 1 integrons continues to be an important source of the genes conferring resistance to the most important antibiotics used clinically, for example imipenem. Most of these resistance genes are found in class 1 integrons, the type that was the subject of the original experimental investigations.
Honours and awards
For her role in the discovery and characterisation of the integron gene cassette system Ruth Hall was elected a Fellow of the Australian Academy of Science in 2005; awarded the American Society of Microbiology Sanofi-Aventis Award for research in the field of Antimicrobial Agents and Chemotherapy also in 2005, was the Australian Society for Microbiology Rubbo Orator in 2004 and received the Australian Society for Microbiology ASM BioMerieux Identifying Resistance Award in 2003.
Source
- Hall RM, 2009, Personal communication.
- Scientist Warns On Antibiotic Resistance, 1998 (Media Release)
- The rise and rise of resistance, 2002 (ECOS Magazine)
