Potyvirus taxonomy
In the late 1980s CSIRO scientists achieved what international experts had believed was impossible – the development of a successful classification system for the largest and most complex group of plant viruses, the Potyviridae. Where previous criteria had failed the CSIRO team showed that coat protein and gene sequence data could be used to classify potyviruses into genera, species and strains, a proposal accepted by the International Committee on Taxonomy of Viruses in 1993.
Their findings on the structure of potyvirus particles explained the long-standing problems in potyvirus serology, led to the generation of virus-resistant plants expressing mouse monoclonal antibodies as well as the development of a novel application in vaccine technology using potyvirus particles as antigen carriers. These particles were highly immunogenic and did not require the use of adjuvants.
The potyvirus problem
The potato virus Y family (potyviruses) is the largest and most complex of the plant virus families, comprising approximately 25% of all known plant viruses. It is estimated to collectively account for more losses to the world’s crop plants than all of the combined effects of all of the other plant virus families. It contains around 200 distinct virus species which in turn account for thousands of strains which collectively cause serious diseases in major agricultural, pasture, horticultural and ornamental crops of commercial significance world-wide as well as in Australia. Potyviruses are currently reported to infect over 2 026 different plant species from 556 genera of 81 different plant families.
In the 1960s, when only a few potyviruses were known, it was relatively easy to establish whether a new isolate causing disease was a distinct virus species or a related strain of a previously known species. However, by the 1980s as more and more potyviruses were discovered with overlapping host ranges and common symptoms, the problem became very difficult. None of the approaches successfully applied to other viruses, such as serology and biological properties, were capable of giving consistent results with the potyvirus family.
A rocky start
Although CSIRO’s potyvirus work was ultimately successful, it was besieged by problems at the start. In 1978, one year after the appointment of plant pathologist Dharma Shukla, the Division of Protein Chemistry was reviewed by an external committee who recommended that the plant virus work should stop. When asked for the Division’s response to this recommendation, Dr Gordon Crewther, the Chief at the time, replied Yes, that’s what they said, but it wasn’t what they meant

Coat proteins provide the answers
In response to the review the emphasis of the project changed. In the period from 1980 to 1993, Dharma Shukla, Colin Ward and their colleagues conducted a major study on the composition and structure of potyvirus particles. Using protein chemical and gene sequencing studies they determined the complete amino acid sequence (or nucleotide sequence) of the coat protein of 16 strains of eight distinct potyvirus species and the partial coat protein sequences of 102 strains of a further nineteen distinct potyvirus species.
This data showed conclusively that the coat protein and gene sequences could be used to distinguish virus species from strains and to establish evolutionary relationships between sets of distinct potyviruses. Comparisons of coat protein amino acid sequences showed that virus strains were much more closely related than distinct virus species. These findings were not consistent with the ‘continuum’ hypothesis proposed to explain the unsatisfactory taxonomy of potyviruses at this time. Their observed bimodal distribution of sequence identity frequencies showed a clear demarcation between potyvirus species and strains.
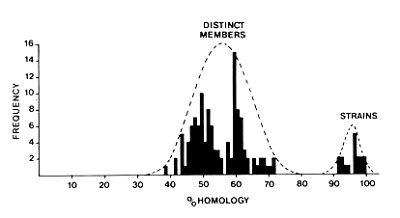
Transforming potyvirus taxonomy
The publication of these findings changed the field. In an extensive series of papers CSIRO scientists applied these new principles to several of the more important potyvirus pathogens. They found that:
- bean common mosaic virus (the most important potyvirus in the USA) consists of two distinct viruses – bean common mosaic virus and bean common mosaic necrosis virus – not a single virus species
- peanut stripe virus (the major pathogen of peanuts world-wide), blackeye cowpea mosaic virus, peanut mild mottle virus, Azuki bean mosaic virus and some strains of cowpea aphid-borne mosaic virus are all strains of bean common mosaic virus
- South African Passiflora virus is not a new distinct virus but a strain of the previously described Morocco strain of cowpea aphid-borne mosaic virus
- pea mosaic virus and white lupin mosaic virus are not distinct species but strains of bean yellow mosaic virus
- the numerous strains assigned to either sugarcane mosaic virus or maize dwarf virus belong to four distinct virus species – sugarcane mosaic virus, maize dwarf mosaic virus, sorghum mosaic virus and Johnsongrass mosaic virus.
The CSIRO team suggested that the aphid- and non-aphid-transmitted potyviruses should be placed into a single family – the Potyviridae – not two distinct families as proposed by others. Further, they suggested that this single family should be divided into genera, species and strains based on their relative sequence identities. Their proposal was approved by the International Committee on Taxonomy of Viruses (ICTV) at its meeting in Glasgow in August 1993. This concept has now been universally accepted by workers and has led to the development of simpler methodologies for rapid virus identification.
Writing to CSIRO in 2006 on the impact of this work Professor Adrian Gibbs, FAA and Life Member of the International Committee on Taxonomy of Viruses (ICTV) stated:
The potyviruses are perhaps the largest of the three largest and most economically important groups of plant viruses – they cause vast amounts of crop damage in all parts of the world, and they are widespread too in wild plants. Despite considerable numbers of studies worldwide in the 1930-80s, they had proved refractory to the traditional methods of characterisation (e.g. serological comparisons of virion proteins, host range studies, etc), and so many had concluded that there might be no clear potyvirus species, just ‘mutant clines’.
Ward and Shukla and their group boldly showed that molecular studies could provide unequivocal evidence that there are discrete potyvirus species that can be clearly defined by gene sequence analysis, and they also revealed the molecular basis of the earlier confusion.
Their work was reported to the world in a large series of very well written landmark publications. These studies are now classics and had a great influence on others. Firstly they opened up the study of potyviruses, so that they are now arguably the best studied group of plant viruses. Secondly the results of their studies, together with those of the very few other groups leading the world in the late 80s-early 90s, made others realise the crucial value of gene sequence comparisons as a basis of almost all modern biological studies, whatever their aim. ‘The rest’, they say, ‘is history’.
Virus particle structure and serology
The team’s findings on the structure of the potyvirus particle and the observation that the highly variable N-terminal region of the potyvirus coat protein was exposed on the surface of the virus particle and was immunodominant explained the molecular basis of the long-standing problems of potyvirus serology. The inconsistent and contradictory serological relationships that dogged the field in the past were shown to be due to variable proportions of antibodies targeted to the conserved core region of the coat protein.
Virus-resistant transgenic plants
These studies on potyvirus serology and particle assembly led to the development of transgenic virus resistant plants which express broadly cross-reactive monoclonal antibodies directed at the conserved coat protein core. This research was done in collaboration with Thomas Joseph Higgins and Paul Chu at CSIRO Plant Industry.
Vaccine technology
The studies on particle assembly and serology also led to a novel application in the field of vaccine technology where the potyvirus coat protein is used as a self-polymerising core scaffold. Fusion proteins with the antigen of choice at the N-terminus were shown to form virus-like particles displaying the foreign antigen on the surface of the particle and to generate strong immune responses in the absence of adjuvant. These virus-like particles carrying vaccine antigens could be produced readily by bacteria, yeast or mammalian cell fermentation.
Recognition
In recognition of their contributions to the identification and classification of potyviruses Colin Ward and Dharma Shukla were awarded the CSIRO Chairman’s Medal in 1994 and the Australian Medal of Agricultural Science in 1995. They were also elected Fellows of the Australian Institute of Agricultural Science in 1995.
Dharma Shukla was Chairman of the International Working Group on Tropical Virology, as well as a member of both the International Working Group on Potyviruses and International Working Group on Legume Viruses.
Colin Ward was elected an Executive Committee Member of the International Committee on the Taxonomy of Viruses (ICTV) from 1996-2002. He was also elected a Fellow of the Australian Academy of Technological Sciences and Engineering in 1998.

Source
- Ward CW, 2009, Personal communication.
